|
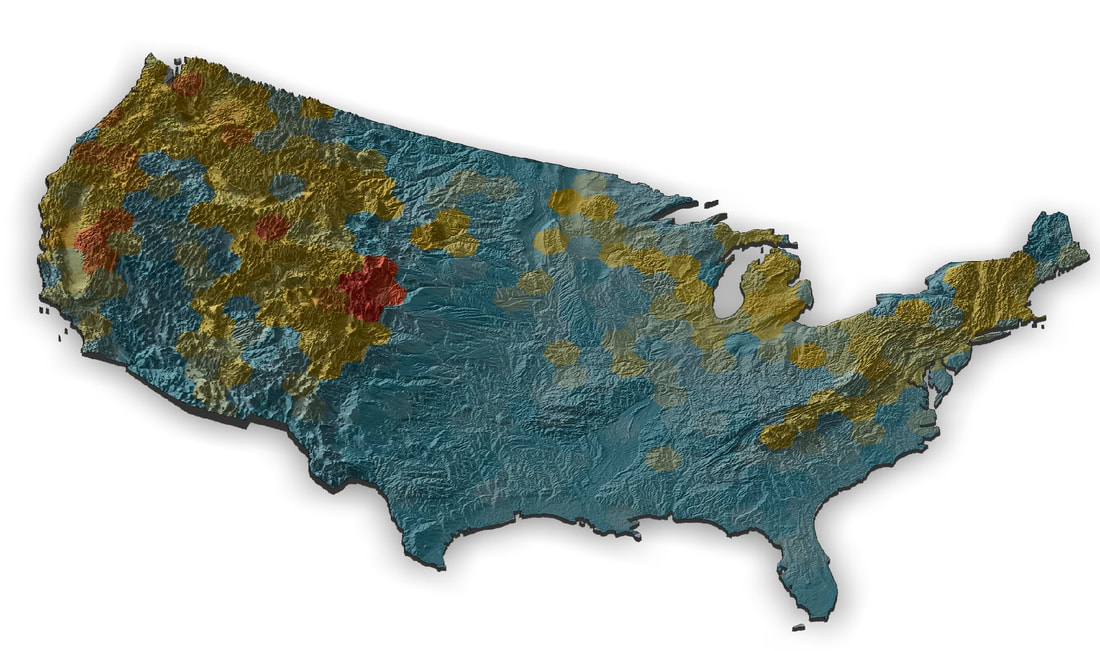
I'm working on a museum display as part of the outreach activities for our NSF Understanding the Rules of Life mountain epibeenomics grant and decided to illustrate why mountains are especially interesting for bumbles by making a map overlaying species richness with elevation. I just used some fairly heavily post-filtered data downloaded from GBIF for this, so could probably be easily improved on with some more rigorous filtering (although the richness blobs correlate pretty well with Paul Williams' maps, so they are pretty good I think). I determined a gridded richness estimate using R with mostly the sf and raster packages and made the pretty map with rayshader. It was a lot of work to wrangle the data and get all the layers in the correct formats, but came out pretty well I think.
PhD student Jamie Bucholz's poster took first place at the 2021 Freshwater Mollusk Conservation Society annual meeting!! Woot. Check it out over on mussels page.
Update, Jamie got a nice plaque.
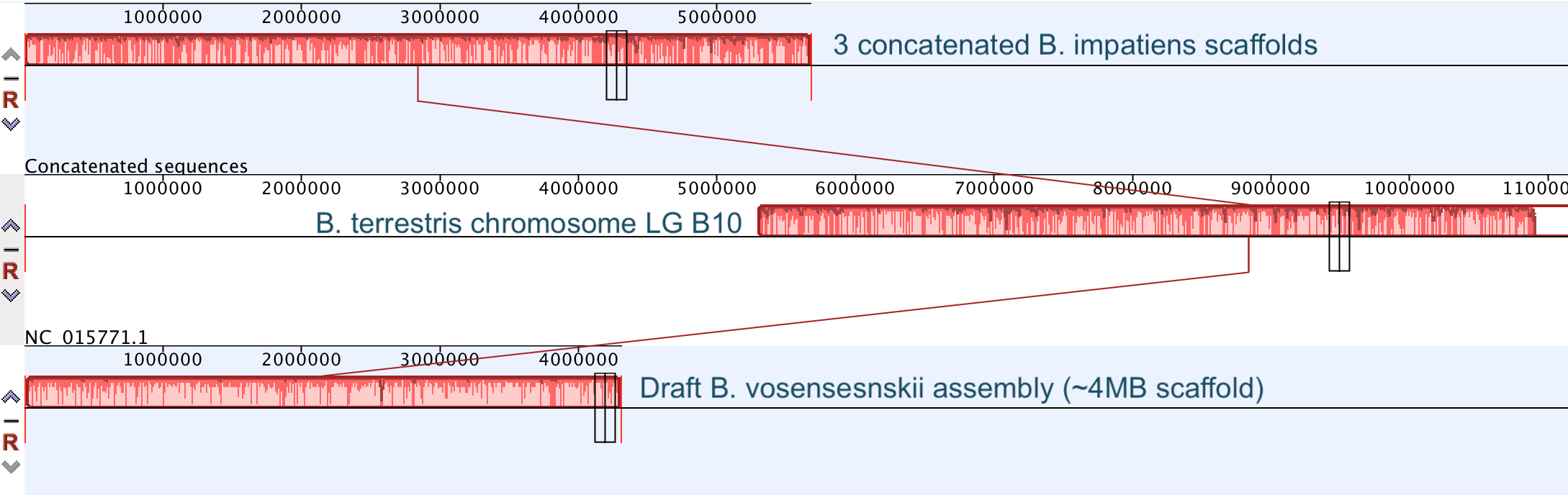
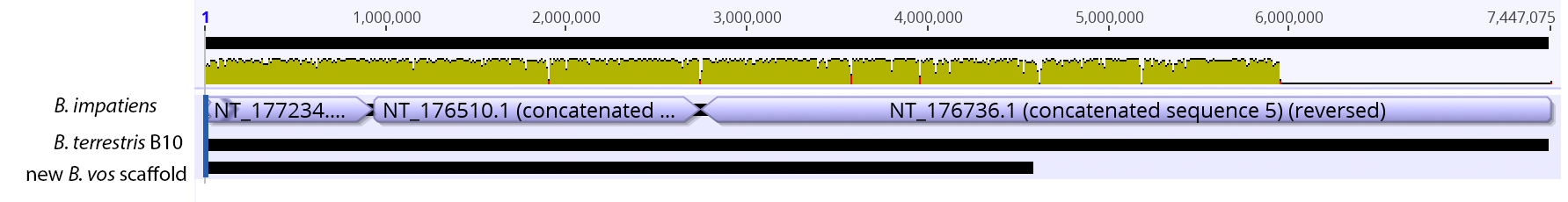
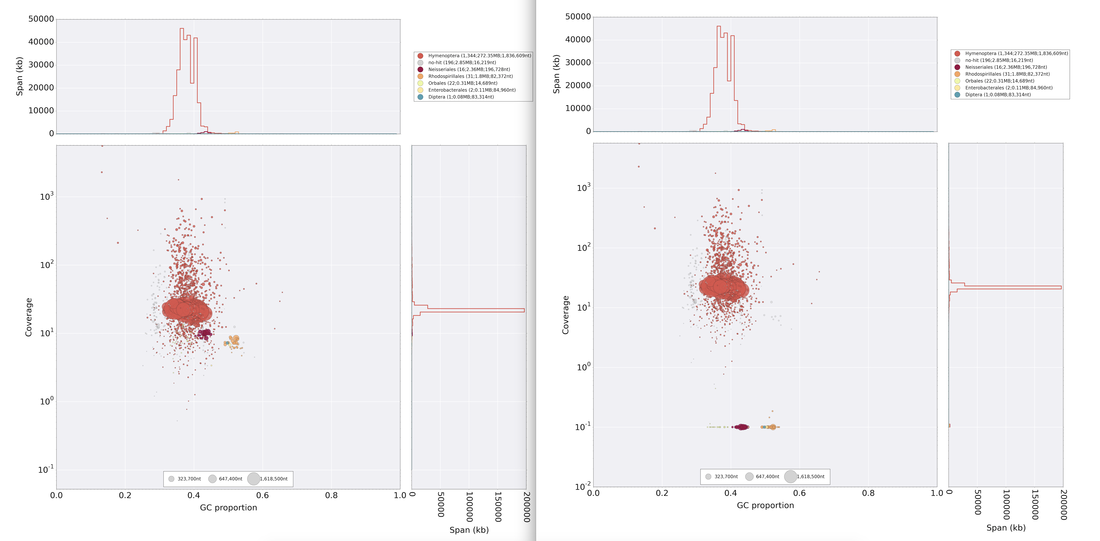
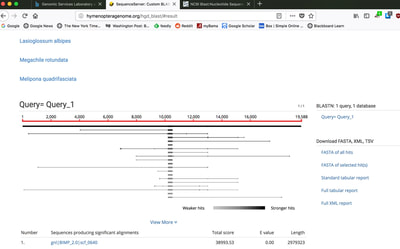
 Excellent news! In collaboration with Carla Atkinson at UA and Colin Jackson and Ryan Garrick at Ole Miss, we have received funding from the NSF Dimensions of Biodiversity Program to investigate biodiversity in freshwater mussels in the southeastern USA, which is THE biodiversity hotspot for these imperiled organisms. The project is highly multi-disciplinary, including population genomics, microbiome, phylogenomics, and functional traits/ecosystem ecology! This is very exciting for us, check out the brand new project website: mussels.ua.edu. We are looking to get lots of people on board, including postdocs, grads, and undergrads, so check out the opportunities page. In addition, in collaboration with Carla and Matt Jenny at UA, we have received funding from UA's Alabama Water Institute to sequence the genomes of several species of freshwater mussels. So we're expanding on the bees, into the world of another set of cool species, the Unionid mussels! So continuing to play with our nanopore/illumina data in the search for a better reference genome for our target bumble bee taxa. Focusing on B. vosnesenskii here, I did a quick and dirty hybrid assembly that included ~30X Oxford Nanopore minion sequencing from 2 males from the same population, combined with ~100X HiSeq data from one of the same males. The assembly was done with the software MaSuRCA, which I gave ~300Gb RAM and 16 threads of computing power on the cluster, producing an assembly in roughly 3 days. So I'm playing with the data now (I'll probably add to this post as I get some other interesting results), but I just did some interesting exploratory analyses of synteny between my assembly and the two published bumble bees (B. impatiens, which is very scaffoldy, and B. terrestris, which has pretty good chromosome/linkage-group level assembly). I took one of my longer draft scaffolds for B. vosnesenskii and tried to see where it fit relative to other Bombus genomes using MAUVE. It appears that synteny is fully conserved for this ~4Mb chunk of genome (see top image), as expected for bumble bee genomes, but there are a few catches. First, this scaffold clearly belongs to the B. terrestris LG B10, which is ~13Mb in size. So clearly the minion+illumina data was't quite good enough to recover the whole chromosome. However, it does much better than the B. impatiens genome. In order to match up my scaffold, I had to stitch together 3 shorter B. impatiens scaffolds (see bottom image). This result looks like it might hold over the rest of the genome, given the distributions of scaffold lengths. So overall the hybrid assembly approach does a pretty darn good job, somewhere in between B. terrestris with its linkage group level assembly and B. impatiens with its 5000-odd scaffolds, without any mate-pair libraries etc. The grand total here = roughly $1,500 and a few days of work (plus the assistance of the Fierst and McKain labs, who got the instrument in the first place!). Of course, it will be much more work to annotate and stitch together the rest of the genome, but that's for a new grad student to work on! One additional thing we are seeing is that there can be a decent amount of contamination in the assembly, which is not necessarily surprising as we had to squish whole bees (including abdomens) to get enough DNA for the Nanopore runs. Numerous scaffolds stemm from bacteria, etc (most actually seem to be known bee symbionts or otherwise bee-associated, so these might be fun to look at downstream). We are playing around with some filtering applications but I think we've settled on the tool "BlobTools". This set of tools lets you map your raw reads (both Nanopore and Illumina) to the reference assembly, generate a BLAST database, and identify the origins of the contaminating sequences relatively quickly. You can then filter out anything you don't like using a combination of coverage, GC content, and taxonomy and redo your assembly. Just to visualize the effectiveness of cleaning the data you can compare a before and after plot (this is nanopore data). Pretty neat. One thing that's nice is that you can clearly see the big scaffolds in the center of the plot are nice and bee-y. Now to redo the assembly.
So in our effort to outdo ourselves in maxing out Oxford nanopore data we ran through a Bombus bifarius worker (black-banded form from the west coast) to get a genome assembly on which to align our low coverage Illumina WGS. We are quite happy with how this instrument is working for us for the long-read sequencing technology (haven't tried pac-bio yet, but we also don't have one of those upstairs) and are getting some nice data that should be sufficient for genome assemblies for B. vosnesenskii and B. bifarius. Just one run of B. vos (~10X coverage) seems to have given us an assembly with contigs with the integrity of the published B. impatiens genome, and we've gone ahead and done a second flowcell and Hiseq run for the same bee to clean things up. The new B. bifarius runs are really nice however, we got roughly 25Gb of sequence and nice long reads. The pic below is a single random read (yup that block of text is one read!) with a single BLAST hit all the way across against the B. impatiens genome with BeeBase...19kb!! Very exciting. We hope to get the B. vosensenskii and B. bifarius genomes put together over the next semester or so.  Thanks to John Sutton from the Fierst lab who is becoming an expert in all things nanopore and has been helping us out with the libraries and monitoring the instrument. Here's a pic of John and I very carefully doing Ampure bead cleanups of the libraries! Jason's first first-authored paper "Distance, elevation, and environment as drivers of diversity and divergence in bumble bees across latitude and altitude" has just been accepted for publication in Molecular Ecology! Congrats all!
Edit (06/04/18) Paper is now up on Molecular Ecology, https://doi.org/10.1111/mec.14735 Now, who didn't wake up this morning and think: "I wonder what it would be like to be a bumble bee?". OK, maybe just me. Nonetheless, now you can find out! As part of our NSF mountain bees grant Broader Impacts, my collaborator Michael Dillon and folks at the U Wyoming Biodiversity Institute (thanks Brian Barber and Kyle Summerfield, especially!) devised "Flight of the Bombus" an especially creative interactive video game that illustrates some of the interesting aspects of insect flight, especially the challenges of flight at altitude. I wrote a small grant to the European Society for Evolutionary Biology and got some additional outreach funds to build a version of the game here. Pretty neat eh! We are not entirely sure where this will go long-term, but we intend to bring it out for various events, take it to local schools etc. A pretty creative endeavor! Go Team!  Lee Roop wrote up a very nice article about our bumble bee research on AL.com. The article also does a nice job of highlighting all the great opportunities for research here at UA. I believe he is planning on doing similar stories about other Alabama scientists as a regular article series, so keep an eye out! Clare and Rebecca have finally finished processing and pinning up all of the bees collected in two bee bowl surveys at the UA Arboretum from last October and November! It looks like we've got a bunch of different species but things were dominated by what I'm guessing from some cursory identification work is the Halictid Agapostemon virescens (my non-bumble bee ID skills are not great, but I'm working on it!), a very pretty metallic green bee. We plan to do another set of monthly surveys this spring to see how diversity changes over the year.
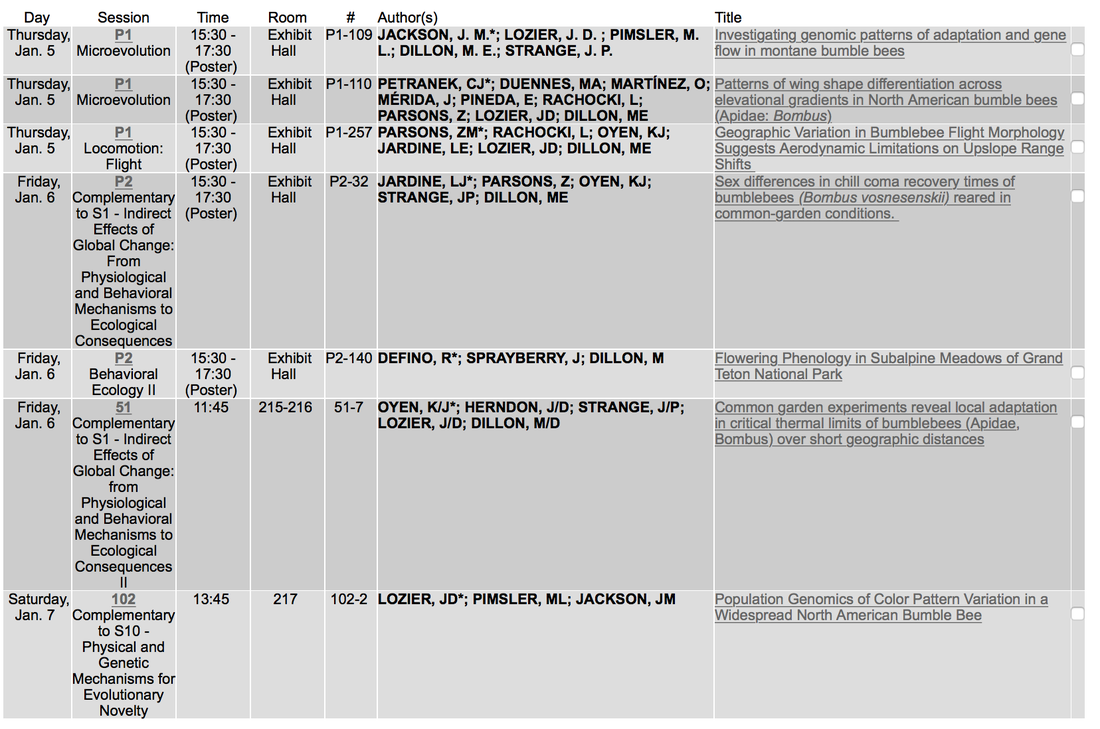
The Lozier and Dillon Labs and our research into local adaptation across montane landscapes will be well represented at the upcoming Society of Integrative and Comparative Biology meeting in New Orleans this January!
 UA Arboretum attracts a crowd at the 2016 Open House UA Arboretum attracts a crowd at the 2016 Open House We are getting ready to undertake a local bee biodiversity inventory at the University of Alabama Arboretum, a beautiful spot off of campus that is undergoing continual improvement thanks to director Monica Watkins. As part of this improvement, we hope to get a baseline assessment of bee biodiversity across the Arboretum, which includes pine and oak/hickory forest, a community garden, wildflower garden, and open areas with lots of general floral and habitat resources that should be great for bees. Ideally, bee habitat availability will increase at the Arboretum over time, and we would like to see how diversity tracks improvements.
 The following is a writeup by Kennan Oyen, a UWY grad student working with collaborator Michael Dillon and describes some of our integrative aspects of the NSF Mountain Bees project. I’m Kennan Oyen, A Ph.D. student in Dr. Michael Dillon’s lab. The Dillon Lab has been busy this summer! Our collaborator Jamie Strange from Utah State and USDA, along with his students, collected wild bumble bee queens (Bombus vosnesenskii) this spring and nested them in the lab. In May, we received our first batch of hives reared from wild queens collected in Southern California. With a team of undergraduates and a high school student I have been studying how these bees cope with extreme temperatures.  We recently wrapped up our first forray into the field for the 2016 season, travelling out west from Alabama to Washington, Oregon, and California. For those interested in what it takes to do bumble bee field work, this picture more or less sums things up. Nets, vials, coolers for storing ice and dry ice to preserve specimens, cooking gear, tents, sleeping bags, and you should be good to go! All this gear does make flying tricky, and since we need a vehicle when we get out west anyway, we usually just drive. Jason and I have driven back and forth across the country together about 4 times now. |
Lozier Lab NewsDispatches from the lab and field! Archives
March 2023
Categories
All
|










 RSS Feed
RSS Feed