|
Congratulations to UA BSC graduate Peter Scott, who went on to do great things as a postdoc out at UCLA, and is now an Assistant Professor at Assistant Professor at West Texas A&M Univ.
Peter just had a first-authored paper in Science (plus the cover!!). Peter studied tortoises that were part of a big relocation project out in southern CA and looked to try to predict what predicted survival of the relocated individuals. The idea is really simple, but provides excellent data that supports a classic assumption that underlies much of modern population/conservation genetics: more genetic variation = greater fitness. This is often difficult to test, but in this case they had the data on individual survival. They use extensive RAD-tag SNP set and look at probability of survival based on individual heterozygosity, translocation distance, and geographic origin... lo and behold, heterozygosity was the key predictor of tortoise survival. Really cool stuff. Congrats Peter! Scott, P. A., Allison, L. J., Field, K. J., Averill-Murray, R. C., & Shaffer, H. B. (2020). Individual heterozygosity predicts translocation success in threatened desert tortoises. Science, 370(6520), 1086–1089. https://doi.org/10.1126/science.abb0421 EntSoc was a little weird this year being all virtual and all, but it seems like the recorded talks were still well attended and the meeting was successful overall (I didn't attend this year because I'm pulling my hair out with regular academic duties...). That also didn't stop Sam Heraghty from giving a great talk on his population genomics work with bumbles. As part of the Mountain Bees Genome Project, Sam has sequenced hundreds of whole genomes from across the ranges of B. vosnesenskii and B. vancouverensis/B. bifarius to look for candidate genes adapted to climate using a landscape genomics approach (to expand on our prior work by Dr Jackson), and it was clearly well-received because he won second place for grad student talks! Papers coming soon!
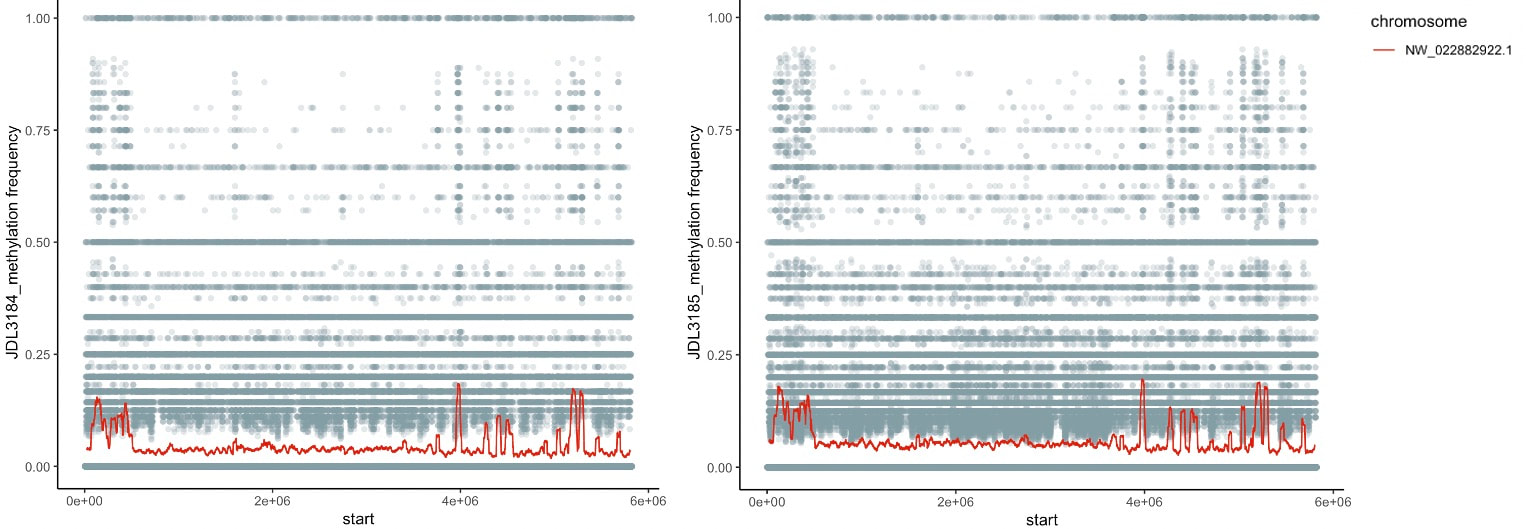
As part of our bee epigenetics NSF project we sequenced some bumble bee genomes, which included Oxford Nanopore Tech. long read sequencing data. We are playing around still, but tried out the pipeline using Nanopolish (available here) to directly identify CpG motifs and identify the fraction of called bases at each motif that appear to be chemically modified. So far I've tried it with the two B. vosnesenskii males (from colonies sourced from same site in OR) we used for the genomes, and see highly consistent results between the individuals across all the scaffods I've peeked at. We have some unpublished bisulfite data for workers from this species, as well as ONT data for other Pyrobombus so will be interesting to compare those data as well.
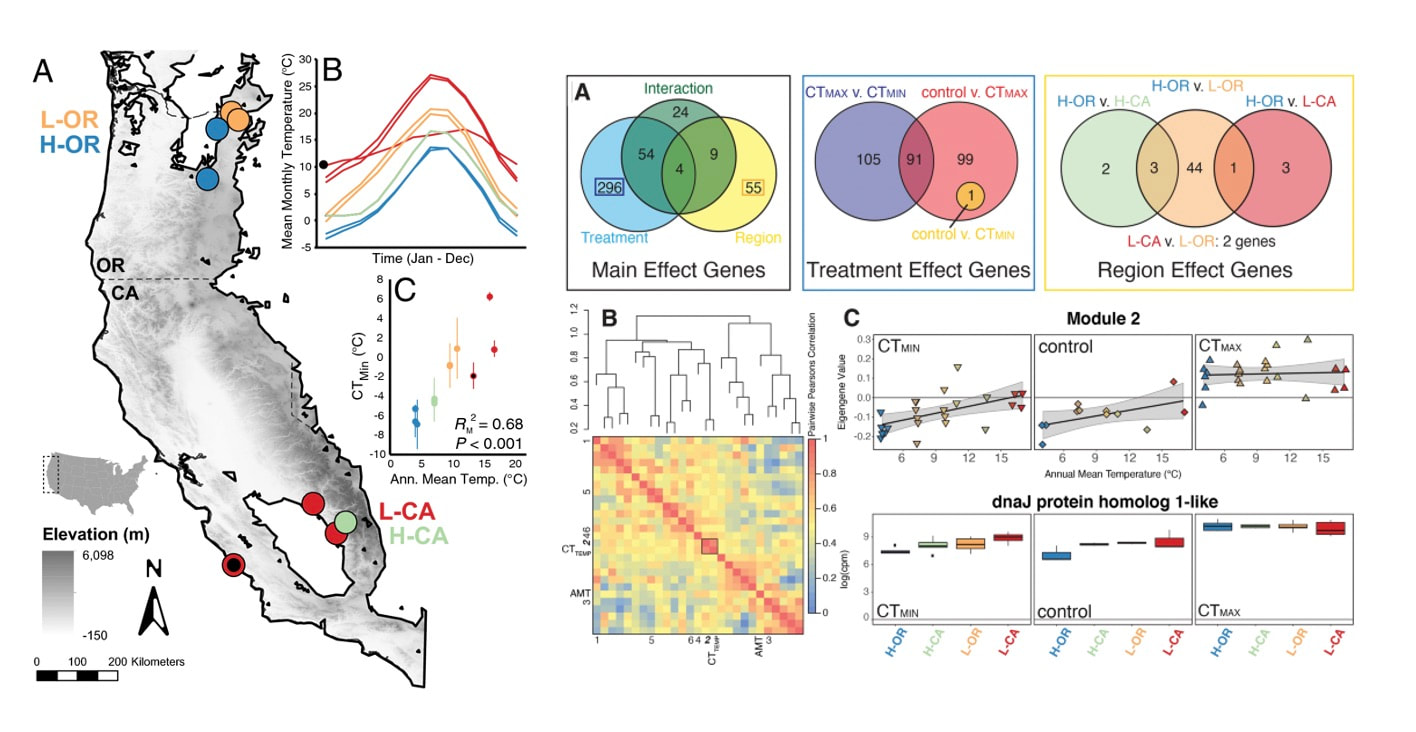
We finally published data from a longstanding project looking at cold adaptation in bumble bees. We started colonies from across the geographic range of B. vosnesenskii (across latitude and altitude) and measured thermal limits (CTmin and CTmax) as well as gene expression in all bees. We found a striking correlation of CTmin with annual mean temperature experienced by each population, and likewise found interesting correlations in gene expression in response to cold across the species range. Intriguingly there was little variation in CTmax or in the genomic response to warming, suggesting species may be constrained in terms of adaptability to increasing environmental temperatures. Pimsler, M.L., Oyen, K.J., Herndon, J.D. et al. Biogeographic parallels in thermal tolerance and gene expression variation under temperature stress in a widespread bumble bee. Sci Rep 10, 17063 (2020). https://doi.org/10.1038/s41598-020-73391-8
|
Lozier Lab NewsDispatches from the lab and field! Archives
March 2023
Categories
All
|

 RSS Feed
RSS Feed