|
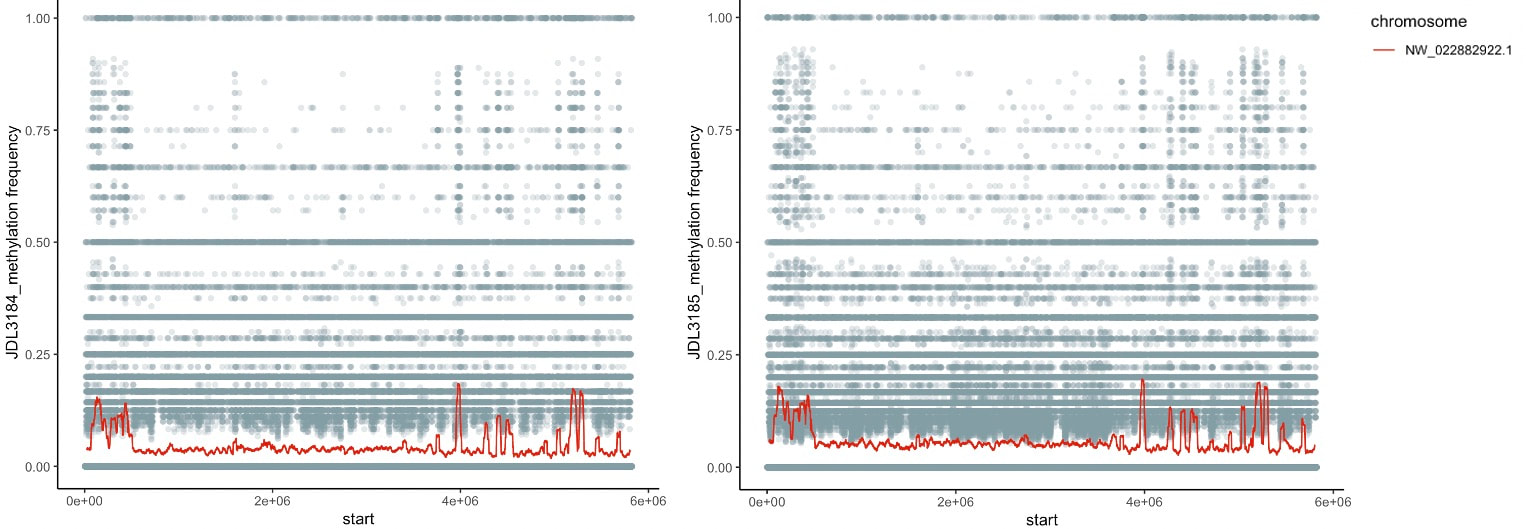
As part of our bee epigenetics NSF project we sequenced some bumble bee genomes, which included Oxford Nanopore Tech. long read sequencing data. We are playing around still, but tried out the pipeline using Nanopolish (available here) to directly identify CpG motifs and identify the fraction of called bases at each motif that appear to be chemically modified. So far I've tried it with the two B. vosnesenskii males (from colonies sourced from same site in OR) we used for the genomes, and see highly consistent results between the individuals across all the scaffods I've peeked at. We have some unpublished bisulfite data for workers from this species, as well as ONT data for other Pyrobombus so will be interesting to compare those data as well.
Comments are closed.
|
Lozier Lab NewsDispatches from the lab and field! Archives
March 2023
Categories
All
|
 RSS Feed
RSS Feed